Research Area: Disease Focus
We develop bioinformatics methods to identify and characterize disease genes and to advance the systems biology understanding of disease processes and the effect of genetic variations on protein function and interactions as well as drug therapies. We devise novel gene prioritization methods using pathway and network data and conduct application studies on the structure, function, and interaction of medically relevant proteins. Another aim is the construction of biological models for the in silico simulation of molecular perturbations underlying disease processes. Most of our work has been performed in close cooperation with research groups from biological and medical institutes associated with NGFN projects on autoinflammatory and neurodegenerative diseases and the two European Networks of Excellence VIRGIL on Hepatitis C and BioSapiens.
Contact: Dr. Mario Albrecht (E-mail: mario.albrecht@mpi-inf.mpg.de)
Further information about Disease Gene Discovery and Autoinflammatory, Neurodegenerative, or Viral Diseases.
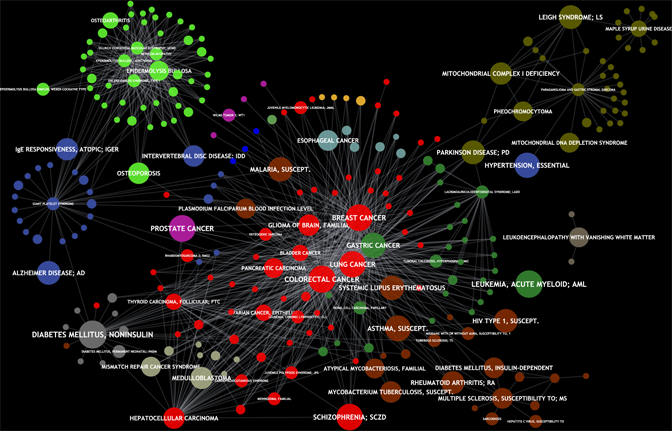
Many diseases are caused by mutations in more than one gene. To discover those disease genes, large-scale linkage and association studies are performed by medical researchers to identify relevant genetic variations (SNPs). However, this approach often yields hundreds of candidate genes that are potentially involved in the disease of interest. Therefore, we develop computational gene prioritization methods that rank putative disease genes to suggest further validating experiments with the most promising ones. The same methods can also be applied to eQTL analyses for finding genes and protein interaction subnetworks underlying certain traits.
Advanced gene identification methods integrate complementary biological information like gene expression data or the knowledge about pathways and protein complexes and consider whole sets of functionally related genes associated with SNPs. Functional similarity has also been shown to be one of the most important features for a successful prioritization method. Therefore, we have extended our FunSimMat web service for comparing disease phenotypes on the molecular level using the Gene Ontology.
In general, FunSimMat is a comprehensive database of precomputed semantic and functional similarity values for the Gene Ontology. It offers several semantic similarity measures for Gene Ontology terms as well as diverse functional similarity measures for all proteins from UniProtKB and for all protein families from Pfam and SMART.
Current Experimental and Medical Cooperation Partners
- Prof. Dr. Ralf Bartenschlager, Department of Molecular Virology, University of Heidelberg, Germany.
- Prof. Dr. Stefan Schreiber, Institute for Clinical Molecular Biology, University of Kiel, Germany.
- Prof. Dr. Thomas F. Meyer, Department of Molecular Biology, Max Planck Institute for Infection Biology, Berlin, Germany.
References
- Doncheva, N.T, Kacprowski, T., Albrecht, M.
Recent approaches to the prioritization of candidate disease genes.
Wiley Interdisciplinary Reviews: Systems Biology and Medicine, 4(5):429-442, 2012.
(Abstract) - Ramírez, F., Lawyer, G., Albrecht, M.
Novel search method for the discovery of functional relationships.
Bioinformatics, 28(2):269-76, 2012.
(Abstract) (Supplement) - Nebot, V., Min, Y., Albrecht, M., Eom, J.H., Weikum, G.
DIDO: a disease-determinants ontology from web sources.
In Proceedings of the 20th International Conference on World Wide Web (WWW 2011), ACM, New York, NY, USA, ISBN 978-1-4503-0637, 237-240, 2011.
(Abstract) (First presented and published at the 20th International World Wide Web Conference (WWW), Hyderabad, India, 2011.) - Schlicker, A., Lengauer, T., Albrecht, M.
Improving disease gene prioritization using the semantic similarity of Gene Ontology terms.
Bioinformatics, 26(18):i561-i567, 2010.
(Abstract) (Supplement) (First presented and published at the 9th European Conference on Computational Biology (ECCB), Ghent, Belgium, 2010.) - Schlicker, A., Albrecht, M.
FunSimMat update: new features for exploring functional similarity.
Nucleic Acids Research, 38(Database issue):D244-248, 2010.
(Abstract) - Schlicker, A., Albrecht, M.
FunSimMat: a comprehensive functional similarity database.
Nucleic Acids Research, 36(Database issue):D434-D439, 2008.
(Abstract)
Non-synonymous SNPs in the homologous gene products NALP3/PYPAF1 and NOD2/CARD15 have been associated with several autoinflammatory diseases that, although clinically different, share a similar inflammatory pathophysiology.
NALP3/PYPAF1 has been linked to chronic infantile neurological cutaneous and articular syndrome (CINCA, also known as neonatal-onset multisystem inflammatory disease, NOMID), familial cold autoinflammatory syndrome (FCAS, also called familial urticaria, FCU), and Muckle-Wells syndrome (MWS). NOD2/CARD15 has been found to confer susceptibility to Blau syndrome (BS, also named arthrocutaneouveal granulomatosis, ACUG) and one of the two main types of chronic inflammatory bowel disease (IBD), Crohn's disease (CD). NALP3 and NOD2 are both involved in the regulation of inflammatory immune responses.
The comparative analysis of the genetic variations with respect to their structural impact on the protein level gives important insights into disease mechanisms (see also this NGFN article). We assembled multiple sequence alignments of the family members sharing a homologous domain architecture with a central NTPase domain called NACHT and a C-terminal LRR (leucine-rich repeat) domain. Our sequence alignments and 3D structural domain models demonstrate that most of the disease-associated variants are located in highly conserved and spatially adjacent regions of the NACHT domain and possibly impair NTP-hydrolysis or oligomerization (Figures 1 and 2).
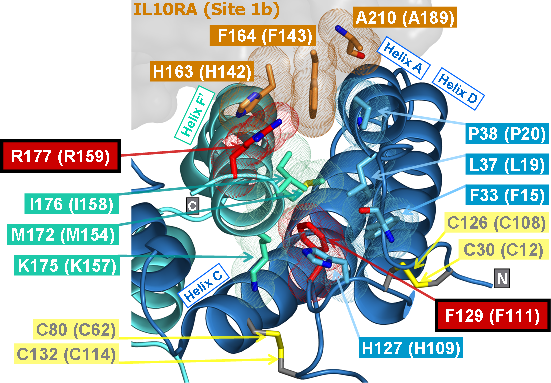
Recently, we could also model the 3D location of IL10 mutations (Figure 3) and contributed to the analysis of the membrane-associated gyanylate kinase DLG5, the autophagy-related 16-like 1 gene ATG16L1, and NELL1, all of which are found associated with IBD. Apart from that, we also studied the 3D structure of interferon-inducible IFI-200 proteins, which are transcriptional regulators involved in inflammation and cancer.
Another structural model of the BTNL2 gene product, a butyrophilin-like member of the immmunoglobulin superfamily, supported experimental investigations on the truncating splice site mutation associated with the multisystemic immune disorder sarcoidosis (Figure 4). We also analyzed the protein structure and function of the epidermal collagen gene COL29A1 associated with atopic dermatitis.
Moreover, we have supported the analysis of plant resistance (R) proteins such as the tomato NBS-LRR protein I-2, which are functionally and structurally related to human NACHT-LRR proteins (NLRs).
Figures 1 and 2 (click on picture for enlargement)
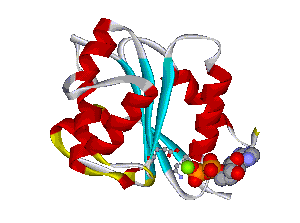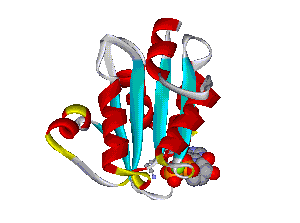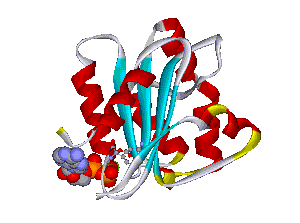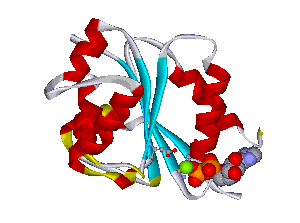
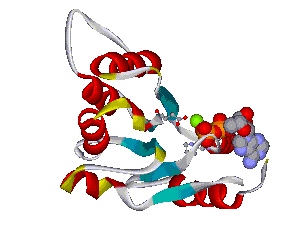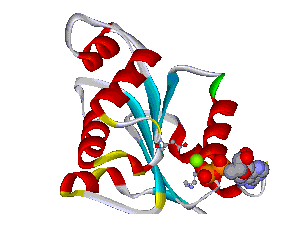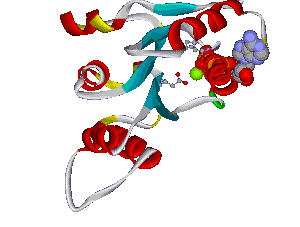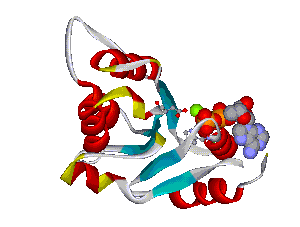
Figures 3 and 4 (click on picture for enlargement)
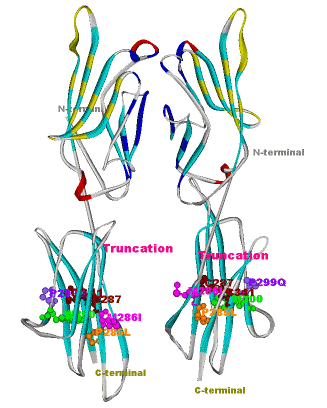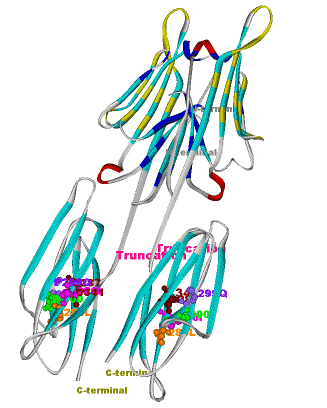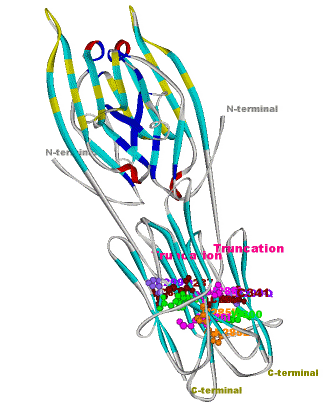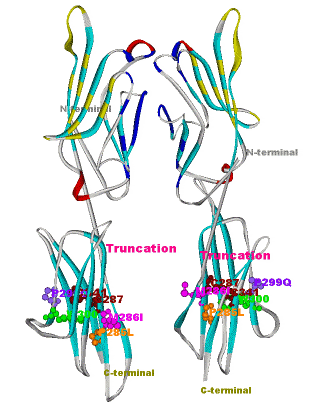
Current Experimental and Medical Cooperation Partners
- Prof. Dr. Stefan Schreiber, Institute for Clinical Molecular Biology, University of Kiel, Germany.
- Dr. Frank Takken, Swammerdam Institute for Life Sciences, University of Amsterdam, The Netherlands.
References
- Ellinghaus, E., Ellinghaus, D., Stuart, P.E., Nair, R.P., Debrus, S., Raelson, J.V.,
Belouchi, M., Fournier, H., Reinhard, C., Ding, J., Li, Y., Tejasvi, T., Gudjonsson, J.,
Stoll, S.W., Voorhees, J.J., Lambert, S., Weidinger, S., Eberlein, B., Kunz, M.,
Rahman, P., Gladman, D.D., Gieger, C., Wichmann, H.E., Karlsen, T.H., Mayr, G., Albrecht, M.,
Kabelitz, D., Mrowietz, U., Abecasis, G.R., Elder, J.T., Schreiber, S., Weichenthal, M., Franke, A.
Genome-wide association study identifies a psoriasis susceptibility locus at TRAF3IP2.
Nature Genetics, 42(11):991-995, 2010.
(Abstract) (Supplement) (Press release) - Franke, A., Balschun, T., Sina, C., Ellinghaus, D., Häsler, R., Mayr, G., Albrecht, M.,
Wittig, M., Buchert, E., Nikolaus, S., Gieger, C., Wichmann, H.E., Sventoraityte, J., Kupcinskas, L.,
Onnie, C.M., Gazouli, M., Anagnou, N.P., Strachan, D., McArdle, W.L., Mathew, C.G., Rutgeerts, P.,
Vermeire, S., Vatn, M.H., the IBSEN study group, Krawczak, M., Rosenstiel, P., Karlsen, T.H., Schreiber, S.
Genome-wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 (IL17REL).
Nature Genetics, 42(4):292-294, 2010.
(Abstract) (Supplement) (Press release) - Brunner, S., Hurni, S., Streckeisen, P., Mayr, G., Albrecht, M., Yahiaoui, N., Keller, B.
Intragenic allele pyramiding combines different specificities of wheat Pm3 resistance alleles.
The Plant Journal, 64(3):433-445, 2010.
(Abstract) (Supplement) - Franke, A., Balschun, T., Karlsen, T.H., Sventoraityte, J., Nikolaus, S., Mayr, G., Domingues, F.S.,
Albrecht, M., Nothnagel, M., Ellinghaus, D., Sina, C., Onnie, C.M., Weersma, R.K., Stokkers, P.C.F.,
Wijmenga, C., Gazouli, M., Strachan, D., McArdle, W.L., Vermeire, S., Rutgeerts, P., Rosenstiel, P.,
Krawczak, M., Vatn, M.H., the IBSEN study group, Mathew, C.G., Schreiber, S.
Sequence variants in IL10, ARPC2, and multiple other loci contribute to ulcerative colitis susceptibility.
Nature Genetics, 40(11):1319-1323, 2008.
(Abstract) (Supplement) (Press release 1) (Press release 2) - Van Ooijen, G., Mayr, G., Kasiem, M.M., Albrecht, M., Cornelissen, B.J., Takken, F.L.
Structure-function analysis of the NB-ARC domain of plant disease resistance proteins.
Journal of Experimental Botany, 59(6):1383-1397, 2008.
(Abstract) (Supplement) - Van Ooijen, G., Mayr, G., Albrecht, M., Cornelissen, B.J., Takken, F.L.
Transcomplementation, but not physical association of the CC-NB-ARC and LRR domains of tomato R protein Mi-1.2 is altered by mutations in the ARC2 subdomain.
Molecular Plant, 1(3):401-410, 2008.
(Abstract) - Hampe, J., Franke, A., Rosenstiel, P., Till, A., Teuber, M., Huse, K., Albrecht, M., Mayr, G.,
De La Vega, F.M., Briggs, J., Günther, S., Prescott, N.J., Onnie, C.M., Häsler, R., Sipos, B.,
Fölsch, U.R., Lengauer, T., Platzer, M., Mathew, C.G., Krawczak, M., Schreiber, S.
A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1.
Nature Genetics, 39(2):207-211, 2007.
(Abstract) (Supplement) - Söderhäll, C., Marenholz, I., Kerscher, T., Rüschendorf, F., Esparza-Gordillo, J., Worm, M.,
Gruber, C., Mayr, G., Albrecht, M., Rohde, K., Schulz, H., Wahn, U., Hubner, N., Lee, Y.A.
Variants in a novel epidermal collagen gene are associated with atopic dermatitis.
PLoS Biology, 5(9):e242.1952-1961, 2007.
(Abstract) (Supplement) (Press release) - Franke, A., Hampe, J., Rosenstiel, P., Becker, C., Wagner, F., Häsler, R., Little, R.D., Huse, K., Ruether, A., Balschun, T., Wittig, M.,
ElSharawy, A., Mayr, G., Albrecht, M., Prescott, N.J., Onnie, C.M., Fournier, H., Keith, T., Radelof, U., Platzer, M., Mathew, C.G.,
Stoll, M., Krawczak, M., Nürnberg, P., Schreiber, S.
Systematic association mapping identifies NELL1 as a novel IBD disease gene.
PLoS ONE, 2(8):e691.1-13, 2007.
(Abstract) (Supplement) - Takken, F.L., Albrecht, M., Tameling, W.I.
Resistance proteins: molecular switches of plant defence.
Current Opinion in Plant Biology, 9(4):383-390, 2006.
(Abstract) (Supplement) - Tameling, W.I., Vossen, J.H., Albrecht, M., Lengauer, T.,
Berden, J.A., Haring, M.A., Cornelissen, B.J., Takken, F.L.
Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation.
Plant Physiology, 140(4):1233-1245, 2006.
(Abstract) - Albrecht, M., Takken, F.L.
Update on the domain architectures of NLRs and R proteins.
Biochemical and Biophysical Research Communications, 339(2):459-462, 2006.
(Abstract) - Schreiber, S., Rosenstiel, P., Albrecht, M., Hampe, J., Krawczak, M.
Genetics of Crohn disease, an archetypal inflammatory barrier disease.
Nature Reviews Genetics, 6(5):376-388, 2005.
(Abstract) - Valentonyte, R., Hampe, J., Huse, K., Rosenstiel, P., Albrecht, M.,
Stenzel, A., Nagy, M., Gaede, K.I., Franke, A., Haesler, R.,
Koch, A., Lengauer, T., Seegert, D., Reiling, N., Ehlers, S.,
Schwinger, E., Platzer, M., Krawczak, M., Muller-Quernheim, J., Schurmann, M.,
Schreiber, S.
Sarcoidosis is associated with a truncating splice site mutation in BTNL2.
Nature Genetics, 37(4):357-364, 2005.
(Abstract) (Supplement) (Press release 1) (Press release 2) - Costello, C.M., Mah, N., Häsler, R., Rosenstiel, P., Waetzig, G.H.,
Hahn, A., Lu, T., Gurbuz, Y., Nikolaus, S., Albrecht, M.,
Hampe, J., Lucius, R., Klöppel, G., Eickhoff, H., Lehrach, H.,
Lengauer, T., Schreiber, S.
Dissection of the inflammatory bowel disease transcriptome using genome-wide cDNA microarrays identifies novel candidate disease genes.
PLoS Medicine, 2(8):e199.1-17, 2005.
(Abstract) (Synopsis) (Press release) - Albrecht, M., Choubey, D., Lengauer, T.
The HIN domain of IFI-200 proteins consists of two OB folds.
Biochemical and Biophysical Research Communications, 327(3):679-687, 2005.
(Abstract) - Van Duist, M.M., Albrecht, M., Podswiadek, M., Giachino, D., Lengauer, T., Punzi, L., De Marchi, M.
A new CARD15 mutation in Blau syndrome
European Journal of Human Genetics, 13(6):742-747, 2005.
(Abstract) - Stoll, M., Corneliussen, B., Costello, C.M., Waetzig, G.H., Mellgard, B.,
Koch, W.A., Rosenstiel, P., Albrecht, M., Croucher, P.J.P., Seegert, D.,
Nikolaus, S., Hampe, J., Lengauer, T., Pierrou, S., Foelsch, U.R.,
Mathew, C.G., Lagerstrom-Fermer, M., Schreiber, S.
Genetic variation in DLG5 is associated with inflammatory bowel disease.
Nature Genetics, 36(5):476-480, 2004.
(Abstract) (Supplement) (Press release) - Albrecht, M., Domingues, F.S., Schreiber, S., Lengauer, T.
Structural localization of disease-associated sequence variations in the NACHT and LRR domains of PYPAF1 and NOD2.
FEBS Letters, 554(3):520-528, 2003.
(Abstract) (Supplement) - Albrecht, M., Lengauer, T., Schreiber, S.
Disease-associated variants in PYPAF1 and NOD2 result in similar alterations of conserved sequence.
Bioinformatics, 19(17):2171-2175, 2003.
(Abstract) - Albrecht, M., Domingues, F.S., Schreiber, S., Lengauer, T.
Identification of mammalian orthologs associates PYPAF5 with distinct functional roles.
FEBS Letters, 538(1-3):173-177, 2003.
(Abstract) (Supplement)
The proteins ataxin-2 and ataxin-3 are the gene products of the spinocerebellar ataxia type 2 and 3 genes SCA2 and SCA3/MJD. Both proteins contain a polyglutamine tract encoded by CAG repeats. Its expansion beyond a certain threshold causes the associated autosomal-dominantly inherited neurodegenerative disorders.
Both diseases SCA2 and SCA3 belong to a heterogeneous group of trinucleotide repeat disorders, which includes Huntington's disease and several other spinocerebellar ataxia types such as SCA1, SCA7 and SCA10. The disorders share common phenotypical features such as the degeneration of specific vulnerable neuron populations and the presence of intracellular aggregations of the mutant proteins in affected neurons.
To provide a rationale for further experiments, we explored the protein architectures of ataxin-2 and ataxin-3 and the protein interaction networks around homologs of ataxin-2 and ataxin-7. Using structure-based multiple sequence alignments of homologous proteins, we investigated domains, sequence motifs, and interaction partners. Our analyses focused on functional amino acids and the construction of three-dimensional models of the RNA-binding Lsm (Like Sm) domain of ataxin-2 and the de-ubiquitinating Josephin domain of ataxin-3. Based on our findings, we proposed further experiments on important functions of ataxin-2 in RNA metabolism and SH3 domain complexes as well as on the binding mode of ataxin-3 to ubiquitin and the multifunctional ATPase p97/VCP/Cdc48. We could also identify the human homologs PATL1 and PATL2 of yeast Pat1 that work together with Sm-like (Lsm) proteins related to ataxin-2.
Apart from polyglutamine proteins, we could recently point to the putative molecular malfunction of the dardarin protein LRRK2/RIP7 in parkinsonism based on a structural analysis of its protein kinase domain.
Current Experimental and Medical Cooperation Partners
-
Prof. Dr. Reinhard Jahn, Department of Neurobiology, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany.
- Dr. J. Douglas Armstrong, Institute for Adaptive and Neural Computation, University of Edinburgh, United Kingdom.
References
- Ramírez, F., Albrecht, M.
Finding scaffold proteins in interactomes.
Trends in Cell Biology, 20(1):2-4, 2010.
(Abstract) (Supplement) - Scheller, N., Resa-Infante, P., De la Luna, S., Galao, R.P., Albrecht, M.,
Kaestner, L., Lipp, P., Lengauer, T., Meyerhans, A., Díez, J.
Identification of PatL1, a human homolog to yeast P body component Pat1.
Biochimica et Biophysica Acta - Molecular Cell Research, 1773(12):1786-1792, 2007.
(Abstract) (Supplement) - Boeddrich, A., Gaumer, S., Haacke, A., Tzvetkov, N., Albrecht, M., Evert, B.O., Müller, E.C.,
Lurz, R., Breuer, P., Schugardt, N., Plaßmann, S., Xu, K., Warrick, J.M., Suopanki, J.,
Wüllner, U., Frank, R., Hartl, U.F., Bonini, N.M., Wanker, E.E.
An arginine/lysine-rich motif is crucial for VCP/p97-mediated modulation of ataxin-3 fibrillogenesis.
The EMBO Journal, 25(7):1547-1558, 2006.
(Abstract) (Supplement) - Ralser, M., Nonhoff, U., Albrecht, M., Lengauer, T., Wanker, E.E., Lehrach, H., Krobitsch, S.
Ataxin-2 and huntingtin interact with endophilin-A complexes to function in plastin-associated pathways.
Human Molecular Genetics, 14(19):2893-2909, 2005.
(Abstract) (Supplement) - Albrecht, M., Huthmacher, C., Tosatto, S.C.E., Lengauer, T.
Decomposing protein networks into domain-domain interactions.
Bioinformatics, 21(Suppl. 2):ii220-ii221, 2005.
(Abstract) (Supplement) - Ralser, M., Albrecht, M., Nonhoff, U., Lengauer, T., Lehrach, H., Krobitsch, S.
An integrative approach to gain insights into the cellular function of human ataxin-2.
Journal of Molecular Biology, 346(1):203-214, 2005.
(Abstract) (Supplement) - Albrecht, M.
LRRK2 mutations and parkinsonism.
The Lancet, 365(9466):1230, 2005.
(Abstract) - Albrecht, M., Golatta, M., Wüllner, U., Lengauer, T.
Structural and functional analysis of ataxin-2 and ataxin-3.
European Journal of Biochemistry, 271(15):3155-3170, 2004.
(Abstract) (Supplement) - Albrecht, M., Lengauer, T.
Novel Sm-like proteins with long C-terminal tails and associated methyltransferases.
FEBS Letters, 569(1-3):18-26, 2004.
(Abstract) (Supplement) - Albrecht, M., Lengauer, T.
Survey on the PABC recognition motif PAM2.
Biochemical and Biophysical Research Communications, 316(1):129-138, 2004.
(Abstract) - Albrecht, M., Hoffmann, D., Evert, B.O., Schmitt, I., Wüllner, U., Lengauer, T.
Structural modeling of ataxin-3 reveals distant homology to adaptins.
Proteins: Structure, Function, and Genetics, 50(2):355-370, 2003.
(Abstract)
Hepatitis C is an important viral disease with about half a million patients in Germany and more than 170 million people affected world-wide. We use and develop bioinformatics methods to analyze sequence variations and the structure and function of HCV gene products as well as their protein interactions with the human host. We are also interested in modeling the membrane association of HCV protein complexes and in simulating the viral perturbations to human protein networks and signaling pathways.
Contacts: Dr. Christoph Welsch (E-mail: christoph.welsch@mpi-inf.mpg.de) and Dr. Mario Albrecht (E-mail: mario.albrecht@mpi-inf.mpg.de)
Current Experimental and Medical Cooperation Partners
- Prof. Dr. Stefan Zeuzem, Department of Internal Medicine 1, Goethe University of Frankfurt, Germany.
- Prof. Dr. Eva Herrmann, Institute for Biostatistics and Mathematical Modeling, Goethe University of Frankfurt, Germany.
- Prof. Dr. Ralf Bartenschlager, Department of Molecular Virology, University of Heidelberg, Germany.
- Prof. Dr. Silvio Tosatto, Department of Biology, University of Padova, Italy.
- Prof. Dr. Jörg Rahnenführer, Department of Statistics, Dortmund University of Technology, Germany.
- Interdisciplinary DFG-funded Clinical Research Group on Hepatitis C, Goethe University of Frankfurt & Saarland University, Germany. [Press release 1] [Press release 2]
A joint BioSapiens-viRgil Workshop on Bioinformatics for Viral Infections was held in Bonn, Germany, on 21-23 September 2005. Press release 1. Press release 2. For more information, please visit http://workshop2005.bioinf.mpi-inf.mpg.de/
References
- Reiss, S., Rebhan, I., Backes, P., Romero-Brey, I., Erfle, H., Matula, P., Kaderali, L., Poenisch, M.,
Blankenburg, H., Hiet, M.S., Longerich, T., Diehl, S., Ramírez, F., Balla, T., Rohr, K., Kaul, A.,
Bühler, S., Pepperkok, R., Lengauer, T., Albrecht, M., Eils, R., Schirmacher, P.,
Lohmann, V., Bartenschlager, R.
Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment.
Host Cell & Microbe, 9(1):32-45, 2011.
(Abstract) (Supplement) (Preview) - Doncheva, N.T., Klein, K., Domingues, F.S., Albrecht, M.
Analyzing and visualizing residue networks of protein structures.
Trends in Biochemical Sciences, 36(4):179-182, 2011.
(Abstract) (Press release) (Press article) - Welker, M.W., Welsch, C., Meyer, A., Antes, I., Albrecht, M., Forestier, N., Kronenberger, B., Lengauer, T., Piiper, A., Zeuzem, S., Sarrazin, C.
Dimerization of the hepatitis C virus nonstructural protein 4B depends on the integrity of an aminoterminal basic leucine zipper.
Protein Science, 19(7):1327-1336, 2010.
(Abstract) - Susser, S., Welsch, C., Wang, Y., Zettler, M., Domingues, F.S., Karey, U., Hughes, E., Ralston, R., Tong, X., Herrmann, E., Zeuzem, S., Sarrazin, C.
Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients.
Hepatology, 50(6):1709-1718, 2009.
(Abstract) (Supplement) - Welsch, C., Zeuzem, S.
RNA-binding activity of hepatitis C virus NS4B: a novel target for small molecule inhibitors.
Gastroenterology, 137(6):2170-2172, 2009.
(Abstract) - Mihm, U., Ackermann, O., Welsch, C., Herrmann, E., Hofmann, W.P., Grigorian, N., Welker, M.W., Lengauer, T., Zeuzem, S., Sarrazin, C.
Clinical relevance of the 2′–5′-oligoadenylate synthetase/RNase L system for treatment response in chronic hepatitis C.
Journal of Hepatology, 50(1):49-58, 2009.
(Abstract) (Supplement) - Mihm, U., Hofmann, W.P., Welsch, C., Polta, A., Lengauer, T., Zeuzem, S., Sarrazin, C., Herrmann, E.
Effect of ribavirin on the frequency of RNase L cleavage sites within the hepatitis C viral genome.
Journal of Viral Hepatitis, 17(3):217-221, 2010.
(Abstract) - Welsch, C., Domingues, F.S., Susser, S., Antes, I., Hartmann, C., Mayr, G., Schlicker, A., Sarrazin, C., Albrecht, M., Zeuzem, S., Lengauer, T.
Molecular basis of telaprevir resistance due to V36 and T54 mutations in the NS3-4A protease of HCV.
Genome Biology, 9(1):R16, 2008.
(Abstract) (Supplement) - Kronenberger, B., Welsch, C., Forestier, N., Zeuzem, S.
Novel hepatitis C drugs in current trials.
Clinics in Liver Disease, 2(3):529-555, 2008.
(Abstract) - Welsch, C., Albrecht, M., Maydt, J., Herrmann, E., Welker, M.W., Sarrazin, C., Scheidig, A., Lengauer, T., Zeuzem, S.
Structural and functional comparison of the non-structural protein 4B in flaviviridae.
Journal of Molecular Graphics and Modelling, 26(2):546-557, 2007.
(Abstract) - Mihm, U., Grigorian, N., Welsch, C., Herrmann, E., Kronenberger, B., Teuber, G., von Wagner, M.,
Hofmann, W.-P., Albrecht, M., Lengauer, T., Zeuzem, S., Sarrazin, C.
Amino acid variations in hepatitis C virus p7 and sensitivity to antiviral combination therapy with amantadine in chronic hepatitis C.
Antiviral Therapy, 11(4):507-519, 2006.
(Abstract) - Hofmann, W.P., Fernandez, B., Herrmann, E., Welsch, C., Mihm, U., Kronenberger, B., Feldmann, G., Spengler, U., Zeuzem, S., Sarrazin, C.
Somatic hypermutation and mRNA expression levels of the BCL-6 gene in patients with hepatitis C virus-associated lymphoproliferative diseases.
Journal of Viral Hepatitis, 14(7):484-491, 2007.
(Abstract) - Welker, M.W., Hofmann, W.P., Welsch, C., von Wagner, M., Herrmann, E., Lengauer, T., Zeuzem, S., Sarrazin, C.
Correlation of amino acid variations within nonstructural 4B protein with initial viral kinetics during interferon-alpha-based therapy in HCV-1b-infected patients.
Journal of Viral Hepatitis, 14(5):338-349, 2007.
(Abstract) - Sarrazin, C., Mihm, U., Herrmann, E., Welsch, C., Albrecht, M., Sarrazin, U., Traver, S., Lengauer, T., Zeuzem, S.
Clinical significance of in vitro replication-enhancing mutations of the hepatitis C virus (HCV) replicon in patients with chronic HCV infection.
The Journal of Infectious Diseases, 192(10):1710-1719, 2005.
(Abstract) - Hofmann, W.P., Herrmann, E., Kronenberger, B., Merkwirth, C., Welsch, C., Lengauer, T., Zeuzem, S., Sarrazin, C.
Association of HCV-related mixed cryoglobulinemia with specific mutational pattern of the HCV E2 protein and CD81 expression on peripheral B lymphocytes.
Blood, 104(4):1228-1229, 2004.
(Abstract) - Kronenberger, B., Sarrazin, C., Hofmann, W.P., von Wagner, M., Herrmann, E., Welsch, C., Elez, R., Ruster, B., Piiper, A., Zeuzem, S.
Mutations in the putative HCV-E2 CD81 binding regions and correlation with cell surface CD81 expression.
Journal of Viral Hepatitis, 11(4):310-318, 2004.
(Abstract) - Zeuzem, S., Welsch, C., Herrmann, E.
Pharmacokinetics of peginterferons.
Seminars in Liver Disease, 23(Suppl. 1):23-28, 2003.
(Abstract)
